Cell Ranger7.1, printed on 04/26/2025
Cell Ranger pipelines run on Linux systems that meet these minimum requirements:
Note: Cell Ranger v6.1 was the last version that supported CentOS/RedHat 6 or Ubuntu 12.04.
The pipelines also run on clusters that meet these additional minimum requirements:
Most software dependencies come bundled in the Cell Ranger package. However, cellranger mkfastq also requires:
| Limit | Recommendation |
|---|---|
| user open files | 16k |
| system max files | 10k per GB RAM available to Cell Ranger |
| user processes | 64 per core available to Cell Ranger |
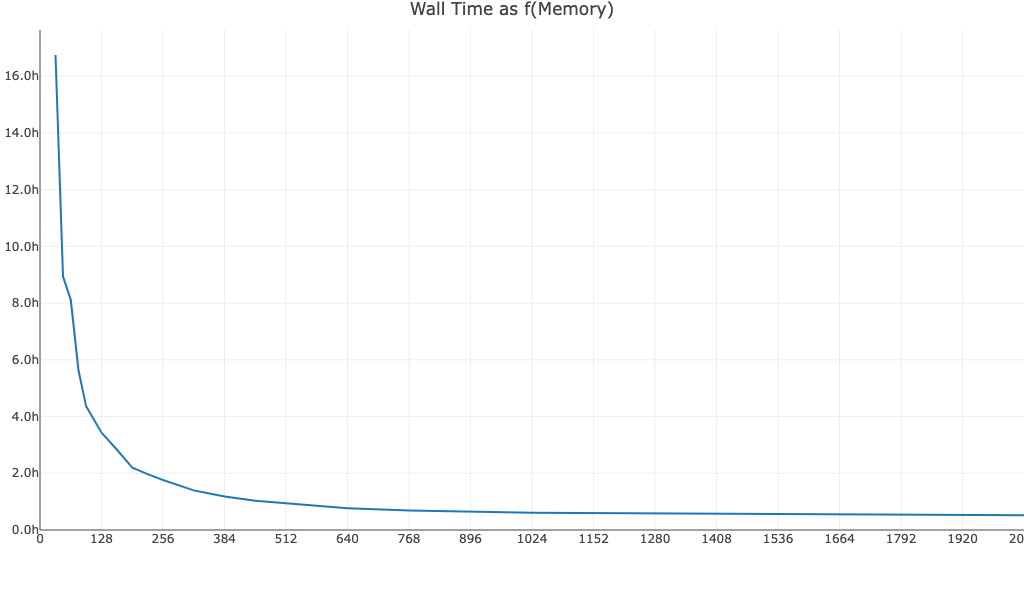
This graph is based on time trials using Amazon EC2 instances. Observed performance will be impacted by other factors besides memory, including read depth, number of cells, number of cores/threads, and I/O. The graph is meant to convey how increasing memory beyond the minimum and recommended levels may impact runtime for large datasets run with cellranger vdj. Similar graphs for cellranger multi, cellranger count, and cellranger aggr are provided on the 3' Single Cell Gene Expression's System Requirements page, but may not be representative of your dataset.
Walltime as a function of memory for a 10k PBMC V(D)J T cell dataset, read depth ~6,500 reads/cell, run with Cell Ranger v6.1.2:
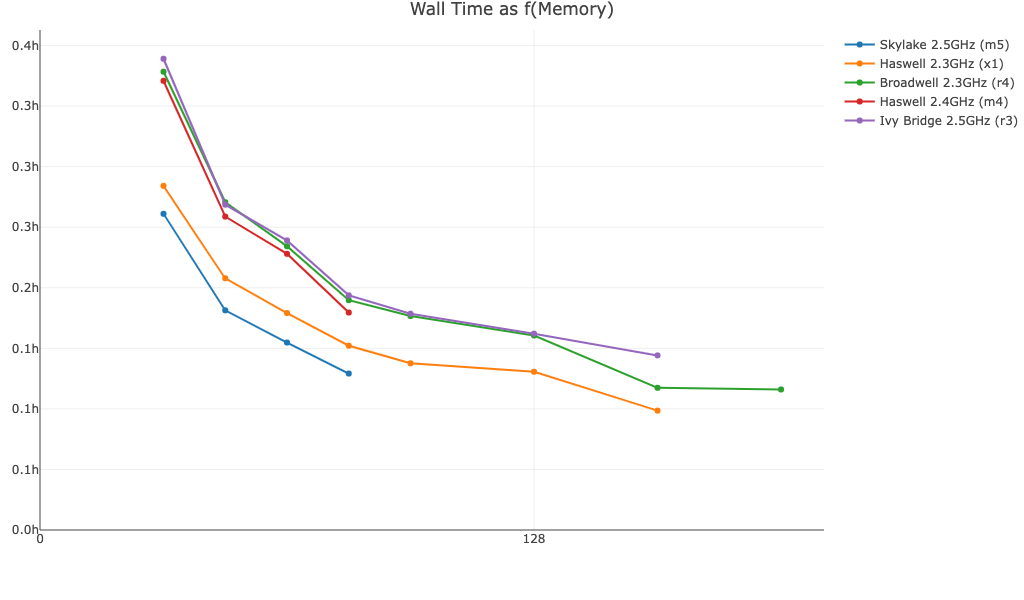
Walltime as a function of memory for a 10k PBMC BCR Antigen Capture dataset run with Cell Ranger v7.1.0:
For similar inputs, walltime as a function of memory is similar for TCR and BCR Antigen Capture datasets.