Space Ranger2.0, printed on 03/03/2025
| After June 30, 2023, new Space Ranger releases will no longer support Targeted Gene Expression analysis. |
Space Ranger provides the same interface and features for both Targeted and Whole Transcriptome Analysis (WTA) Gene Expression data, as described in Single-Library Analysis with spaceranger count. The workflow diagram shows all the required inputs and the corresponding spaceranger count flags for Targeted Gene Expression analysis:
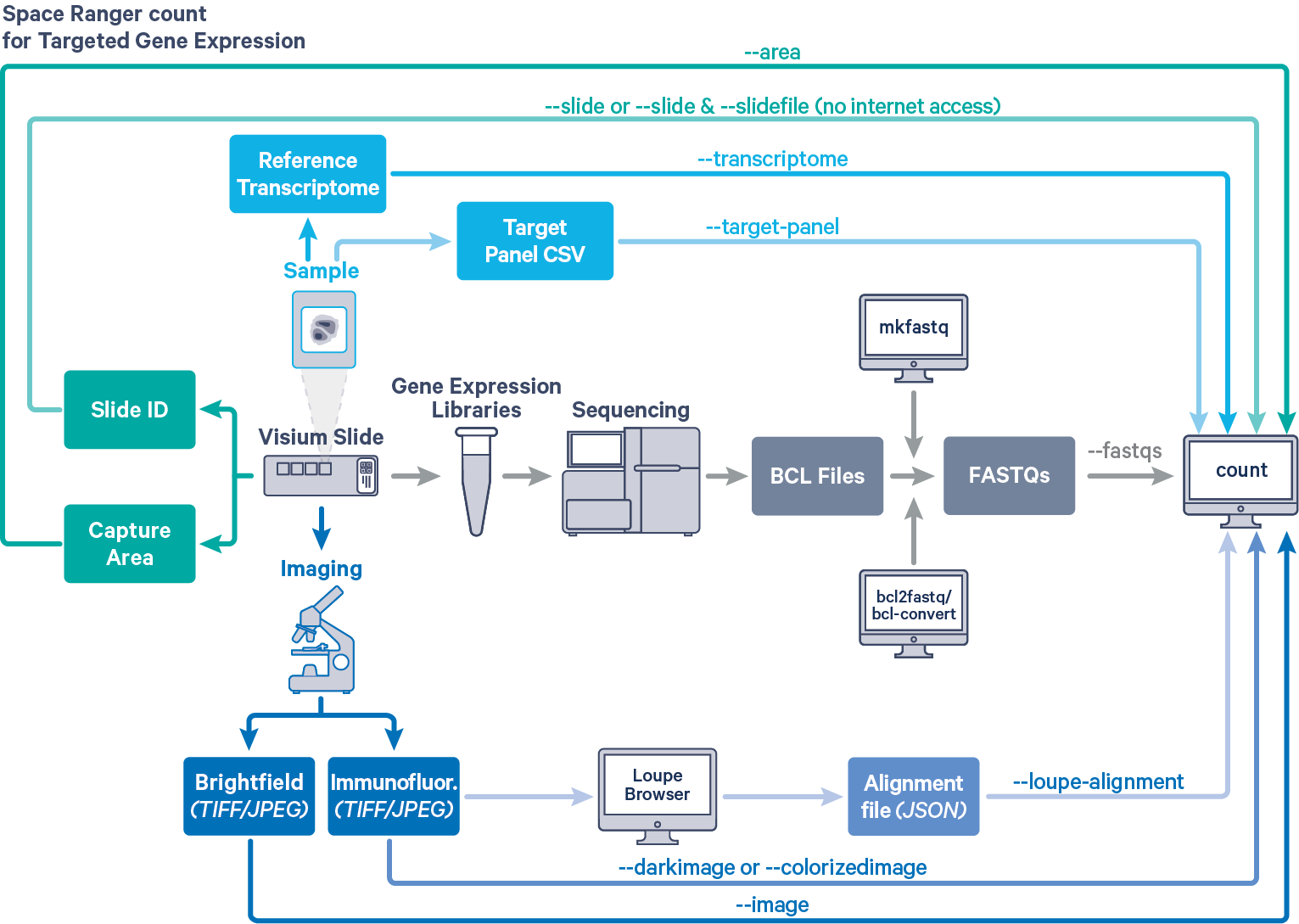
The spaceranger count is run on each individual Capture Area in the Visium slide. The required inputs are:
--fastqs)
TIFF, QPTIFFor JPEG format:
--image for brightfield image--darkimage for dark background fluorescence image--colorizedimage for composite colored fluorescence image--slide & --area if spaceranger has access to internet--slidefile, --slide & --area if spaceranger has no access to internet. The slide layout file is directly downloaded--unknown-slide if visium slide details are unknown--transcriptome)--target-panel flagFor a list of accepted arguments, see the Command Line Argument Reference below, or run spaceranger count --help.
Select the tab corresponding to the imaging workflow.
To generate spatial feature counts for a single library using automatic fiducial alignment and tissue detection on a brightfield image input, run spaceranger count with the following arguments. The code in red indicates user specific inputs.
$ cd /home/jdoe/runs $ spaceranger count --id=sample345 \ #Output directory --transcriptome=/home/jdoe/refdata/GRCh38-2020-A \ #Path to Reference --target-panel=/home/jdoe/spaceranger-1.3.0/target_panels/immunology_v1.0_GRCh38-2020-A.target_panel.csv \ #Path to the Target Panel CSV --fastqs=/home/jdoe/runs/HAWT7ADXX/outs/fastq_path \ #Path to FASTQs --sample=mysample \ #Sample name from FASTQ filename --image=/home/jdoe/runs/images/sample345TIFF \ #Path to brightfield image input --slide=V19J01-123 \ #Slide ID --area=A1 \ #Capture area --localcores=8 \ #Allowed cores in localmode --localmem=64 #Allowed memory (GB) in localmode
To generate spatial feature counts for a single library using a fiducial alignment and tissue assignment
JSON file generated in Loupe Browser on a brightfield image input, run spaceranger count with the following arguments. The code in red indicates user specific inputs.
$ cd /home/jdoe/runs $ spaceranger count --id=sample345 \ #Output directory --transcriptome=/home/jdoe/refdata/GRCh38-2020-A \ #Path to Reference --target-panel=/home/jdoe/spaceranger-1.3.0/target_panels/immunology_v1.0_GRCh38-2020-A.target_panel.csv \ #Path to the Target Panel CSV --fastqs=/home/jdoe/runs/HAWT7ADXX/outs/fastq_path \ #Path to FASTQs --sample=mysample \ #Sample name from FASTQ filename --image=/home/jdoe/runs/images/sample345TIFF \ #Path to brightfield image input --slide=V19J01-123 \ #Slide ID --area=A1 \ #Capture area --loupe-alignment=sample345.json \ #Manual alignment file --localcores=8 \ #Allowed cores in localmode --localmem=64 #Allowed memory (GB) in localmodeThe arguments to specify the fluorescence image can be either
--darkimage or --colorizedimage, depending on the image format. Details about the different supported fluorescence image formats are described in Image Recommendations.
Target panel CSV files corresponding to predesigned gene panels can be found within the target_panels directory in the Space Ranger package. Spatial Gene Expression does not include support for fully custom panels.
target_panels/ ├── gene_signature_v1.0_GRCh38-2020-A.target_panel.csv ├── immunology_v1.0_GRCh38-2020-A.target_panel.csv ├── neuroscience_v1.0_GRCh38-2020-A.target_panel.csv └── pan_cancer_v1.0_GRCh38-2020-A.target_panel.csv
For example, if you installed Space Ranger to /home/jdoe/spaceranger-2.0.0, then the target panel CSVs will be contained in /home/jdoe/spaceranger-2.0.0/target_panels.
For the target panel CSV files, go to:
The Panel Selection page for information about each panel as well as direct downloads of all the accompanying files.
The Panel File Descriptions page for detailed documentation on all the file formats used in targeted analysis.
The output of the pipeline is contained in a folder named with the sample ID you specified (such as sample345). If this folder already exists, in a rerun with the original parameters, spaceranger will assume it is an existing pipestance and attempt to resume running it. The subfolder named outs contains the main pipeline outputs.
The list of all the relevant arguments are described in Single-Library Analysis with spaceranger count or run spaceranger count --help.
| Argument | Description |
|---|---|
--target-panel | Required for Targeted Gene Expression analysis. Path to a Target Panel CSV file declaring the target panel used. |
--rps-limit | Optional for Targeted Gene Expression analysis. Subsample to at most N mean reads under tissue per spot for targeted gene expression if N > 0, or disable subsampling if N = 0. Modifying this parameter is not recommended. The default value of N is 15,000 reads under tissue per spot for targeted gene expression. See Targeted Algorithms for details. |