Space Ranger, printed on 03/03/2025
Loupe Browser 4.0 has been updated to allow users to easily analyze data
generated with the Visium Spatial Gene Expression Solution, while still allowing
users to easily explore single-cell gene expression, immunology, and ATAC data.
It is compatible with Space Ranger 1.0, and remains compatible with .cloupe
files generated by all versions of Cell Ranger after v1.3.
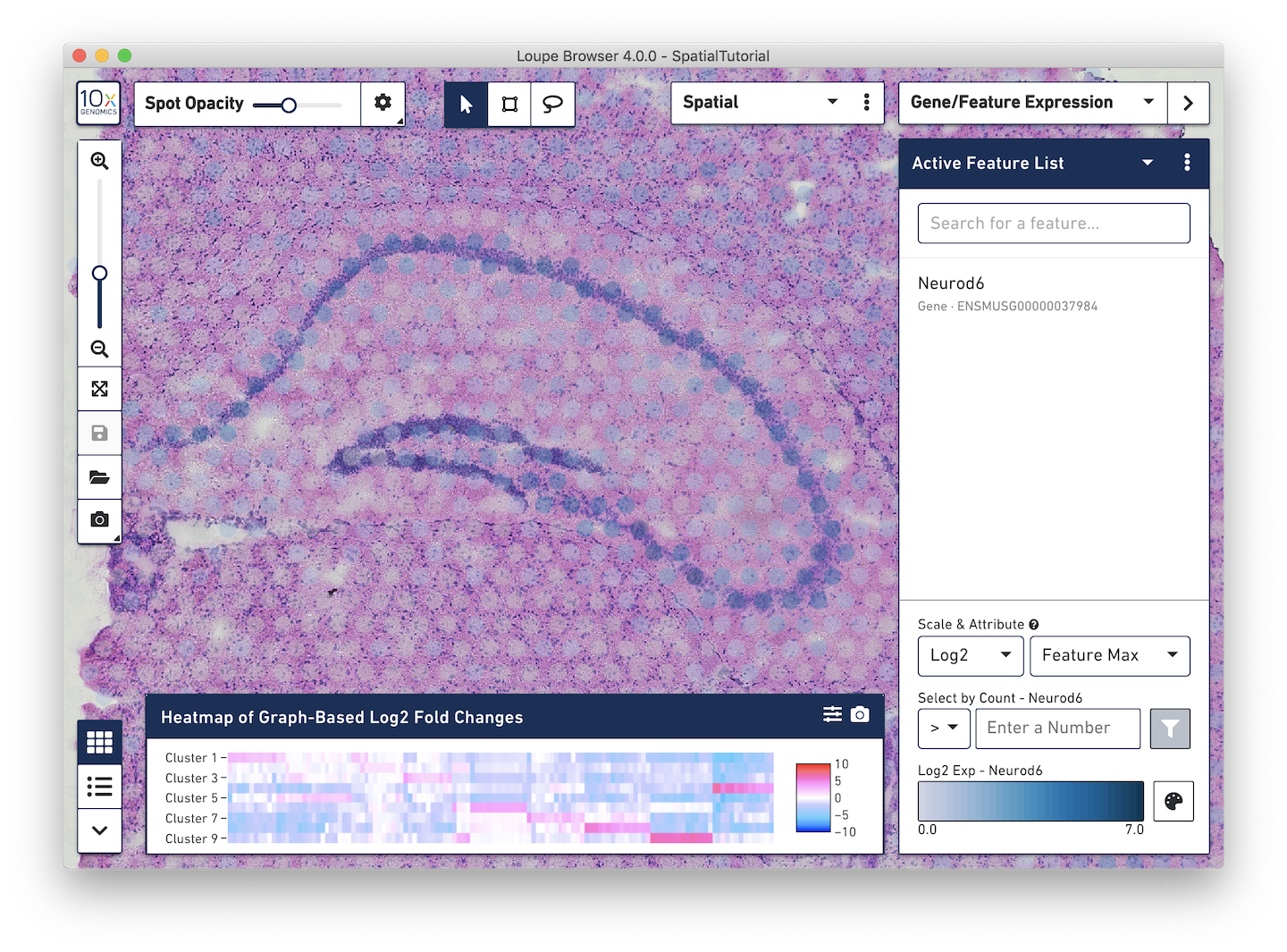
Loupe Browser 4.0 allows users to analyze spatial gene expression data within the context of a high-resolution microscope image:

Loupe Browser's fluid, responsive user interface allows users to quickly label morphological structure, explore the relation between underlying tissue structure and gene expression and identify regions of the tissue with unique gene expression. A variety of graphical settings and a wider range of color palettes will allow users to export the ideal graphics for their experiment. Finally, Loupe Browser 4.0 comes preloaded with a mouse brain tutorial, so existing single-cell users can explore a spatial gene expression dataset.
To find out how to use Loupe Browser 4.0 to analyze spatial gene expression, please consult the Loupe Browser documentation.
Though Space Ranger comes with reliable automatic image detection algorithms, Loupe Browser 4.0 can also be used to override the pipeline's decisions, or provide finer-grained control over tissue labeling for diverse samples.

The Visium Manual Alignment Wizard allows users to easily line up their microscope image with a slide layout file, and to precisely label tissue and background spots for processing by the Space Ranger pipeline.
Enjoy a variety of new color palettes for feature expression, including inverted color scales, Turbo, Viridis and Plasma. The heatmap color scheme should also make it easier to see differences in differential gene patterns between clusters.
Loupe Browser will warn, but not prevent you from doing the following: