Space Ranger6.4, printed on 03/03/2025
Loupe Browser tutorials review the major analysis capabilities available for spatial datasets.
Follow the instruction on the Installation page to download and install OS specific software (Windows or macOS).
Click on the SpatialTutorial.cloupe file from the list of Recent Files. The file opens in a new window with the mouse brain image and the spots
overlayed on top of the image. The spots are color-coded by the cluster that the
spot was assigned to.
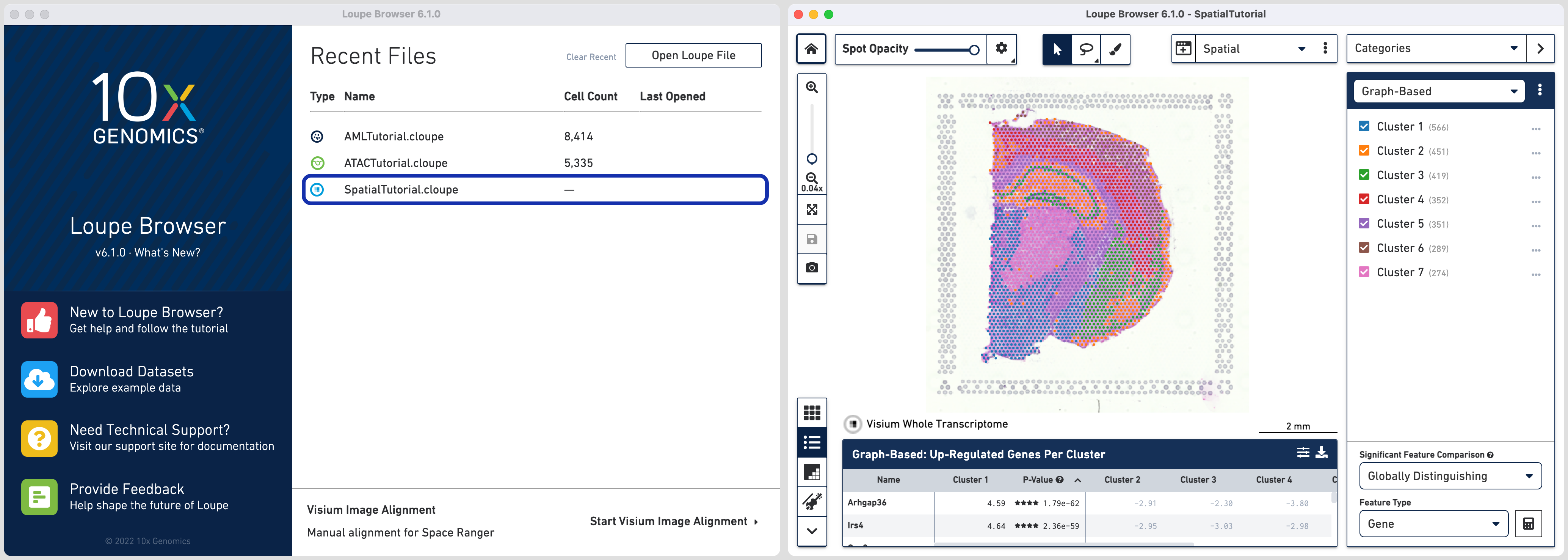
The SpatialTutorial was generated from fresh frozen mouse brain tissue section. A brightfield image of the hematoxylin & eosin (H&E) stained tissue section was acquired. Refer to the dataset page for more about the imaging and sequencing details.
From Loupe Browser v6.1 onwards, the embedded .cloupe file was generated by Space Ranger 1.3 pipeline which was run with automatic fiducial detection and image alignment arguments. The embedded tutorial in older versions of Loupe Browser was generated by Space Ranger 1.0.
There are a number of other Spatial Gene Expression datasets publicly available for download. These datasets include the .cloupe file that you can use to visualize the results. Visit the Spatial Gene Expression datasets page.
Starting with Loupe Browser v6.1, users may choose to share anonymized non-biological usage data (i.e. no identifiable sample information such as files, cluster names, or any open text fields are tracked) to improve Loupe Browser performance. Sharing of usage data is optional, and users can change whether they want to share usage data any time. The user preference for usage data sharing will carry over within a major version (e.g. all 6.Y versions) but the user will be asked to confirm their preferences for each major version update.
In case you wish to change usage data sharing setting, click the Manage Usage Data from the Help menu and change the preference in the pop-up window.