Cell Ranger6.1, printed on 04/16/2025
Cell Ranger 6.0 and later supports analyzing 3' Cell Multiplexing data with the cellranger multi pipeline.
| Coming Soon: the next release of Cell Ranger will count intronic reads by default for whole transcriptome gene expression data. For more information, see our recommendation on including introns for gene expression analysis page. |
The cellranger multi pipeline is required to analyze 3' Cell Multiplexing data. Otherwise, users can continue to use cellranger count.
| 3' GEX | 3' FB | CellPlex | Use multi? |
|---|---|---|---|
| Yes | Yes | Yes | Required |
| Yes | Yes | No | Optional. Prefer count |
| Yes | No | Yes | Required |
| Yes | No | No | Optional. Prefer count |
| No | Yes | No | Optional. Prefer count |
| The Cell Ranger multi pipeline supports the analysis of cell multiplexed data (e.g., CellPlex). While custom tags are not supported by 10x Genomics, Cell Ranger is capable of analyzing cell multiplexed data using custom tags (such as TotalSeqA/B/C). Please see the CMO Reference section for more details. |
First, follow the instructions on running cellranger mkfastq to generate FASTQ files. For example, if the flow cell ID was HAWT7ADXX, then cellranger mkfastq will output FASTQ files in HAWT7ADXX/outs/fastq_path. If you are already starting with FASTQ files, you can skip this step and proceed directly to run cellranger multi.
Running cellranger multi requires a config CSV, described below, invoking the following arguments:
| Argument | Description |
|---|---|
--id | A unique run ID string: e.g. sample345 that is also the output folder name. Cannot be more than 64 characters. |
--csv | Path to config CSV file with input libraries and analysis parameters. |
The multi config CSV contains both the library definitions and experimental design variables. It is composed of up to four sections for 3' data: [gene-expression], [libraries], [feature], and [samples]. Example formats for different product configurations are below.
The [gene-expression] and [feature] sections have at most two columns (Field name and value), and are responsible for configuring their respective portions of the experiment. The [libraries] section specifies where input FASTQ files may be found and has at least three columns. The [samples] section specifies sample information for Cell Multiplexing and has at least two columns.
A template for a multi config CSV can be downloaded here and example multi config CSVs can be downloaded from 6.0 public datasets here.
Starting with Cell Ranger 6.1, it is generally recommended to specify the estimated number of cells in the expect-cells field (defaults to 3,000). If need be, automated cell calling can be overridden with the force-cells field.
|
| Multi Config CSV | |
|---|---|
Section: [gene-expression] | |
| Field | Description |
reference
| Path of folder containing 10x Genomics-compatible reference. Required for gene expression and Feature Barcode libraries. |
min-assignment-confidence
| Optional. Introduced in Cell Ranger 6.0.2. The minimum estimated likelihood to call a sample as tagged with a Cell Multiplexing Oligo (CMO) instead of "Unassigned". Users may wish to tolerate a higher rate of mis-assignment in order to obtain more singlets to include in their analysis, or a lower rate of mis-assignment at the cost of obtaining fewer singlets. By default, this value is 0.9. Contact support@10xgenomics.com for further advice. |
cmo-set
| Optional. CMO set CSV file, declaring CMO constructs and associated barcodes. see CMO Reference section for more detail. |
target-panel
| Optional. Path to a target panel CSV file or name of a 10x Genomics fixed gene panel (pathway, pan-cancer, immunology, neuroscience). |
no-target-umi-filter
| Optional. Disable targeted UMI filtering stage. See Targeted Algorithms for details. Default: false. |
r1-length
| Optional. Hard trim the input Read 1 of gene expression libraries to this length before analysis. Default: do not trim Read 1. |
r2-length
| Optional. Hard trim the input Read 2 of gene expression libraries to this length before analysis. Default: do not trim Read 2. |
chemistry
| Optional. Assay configuration. NOTE: by default, the assay configuration is detected automatically (recommended mode). Users will typically not need to specify a chemistry. Options are: 'auto' for autodetection, 'threeprime' for Single Cell 3', 'SC3Pv1' or 'SC3Pv2' or 'SC3Pv3' for Single Cell 3' v1/v2/v3, 'SC3Pv3HT' for Single Cell 3' v3.1 HT, 'SC-FB' for Single Cell Antibody-only 3' v2. Default: auto. |
expect-cells
| Optional, recommended. Expected number of recovered cells. Default: 3000. |
force-cells
| Optional. Force pipeline to use this number of cells, bypassing cell detection. Default: detect cells using EmptyDrops. |
include-introns
| Optional. Include intronic reads in count. Default: false |
no-secondary
| Optional. Disable secondary analysis, e.g. clustering. Default: false. |
no-bam
| Optional. Do not generate a bam file. Default: false. |
Section: [feature] | |
| Field | Description |
reference
| Feature reference CSV file, declaring Feature Barcode constructs and associated barcodes. Required for Feature Barcode libraries, otherwise optional. |
r1-length
| Optional. Hard trim the input Read 1 of Feature Barcode libraries to this length before analysis. Default: do not trim Read 1. |
r2-length
| Optional. Hard trim the input Read 2 of Feature Barcode libraries to this length before analysis. Default: do not trim Read 2. |
Section: [libraries] (see also Specifying Input FASTQ Files for cellranger multi) | |
| Column | Description |
fastq_id
| Required. The Illumina sample name to analyze. This will be as specified in the sample sheet supplied to mkfastq or bcl2fastq. |
fastqs
| Required. The folder containing the FASTQ files to be analyzed. Generally, this will be the fastq_path folder generated by cellranger mkfastq. |
lanes
| Optional. The lanes associated with this sample, separated by |. Defaults to using all lanes. |
physical_library_id
| Optional. Library type. NOTE: by default, the library type is detected automatically based on specified feature_types (recommended mode). Users typically do not need to include the physical_library_id column in the CSV file. |
feature_types
| Required. The underlying feature type of the library, which must be one of ‘Gene Expression’, ‘Antibody Capture’, ‘CRISPR Guide Capture’, or ‘Multiplexing Capture’. |
subsample_rate
| Optional. The rate at which reads from the provided FASTQ files are sampled. Must be strictly greater than 0 and less than or equal to 1. |
Section: [samples] | |
| Column | Description |
sample_id
| A name to identify a multiplexed sample. Must be alphanumeric with hyphens and/or underscores, and less than 64 characters. Required for cell multiplexing libraries. |
cmo_ids
| The cell multiplexing oligo IDs used to multiplex this sample. If multiple CMOs were used per sample, separate IDs with a pipe (e.g., CMO301|CMO302). Required for cell multiplexing libraries. |
description
| Optional. A description for the sample. |
For help on how to configure the [libraries] section to target a particular set of FASTQs, consult Specifying Input FASTQ Files for cellranger multi.
|
cd /home/jdoe/runs cellranger multi --id=sample345 --csv=/home/jdoe/sample345.csv
Martian Runtime - v4.0.6 Running preflight checks (please wait)... ...
By default, cellranger will use all of the cores available on your
system to execute pipeline stages. You can specify a different number of cores
to use with the --localcores option; for example, --localcores=16
will limit cellranger to using up to sixteen cores at once. Similarly,
--localmem will restrict the amount of memory (in GB) used by
cellranger.
The pipeline will create a new folder named with the run ID you specified using the --id argument (e.g. /home/jdoe/runs/sample345) for its output. If this folder already exists, cellranger will assume it is an existing pipestance and attempt to resume running it.
Waiting 6 seconds for UI to do final refresh. Pipestance completed successfully! yyyy-mm-dd hh:mm:ss Shutting down. Saving pipestance info to "tiny/tiny.mri.tgz"
sample345). The subfolder named outs will contain the main pipeline outputs.Here are a few example multi config CSVs for some common product configurations, along with simplified diagrams for the corresponding experimental set up. Make sure to replace /path/to with the actual full path to your data, and edit any text in red according to the experiment's sample/library/file names. The expect-cells option is recommended; up to 30,000 cells are supported for Cell Multiplexing.
| Experimental Design | Multi config CSV |
3' GEX with Cell Multiplexing, 1 CMO/sample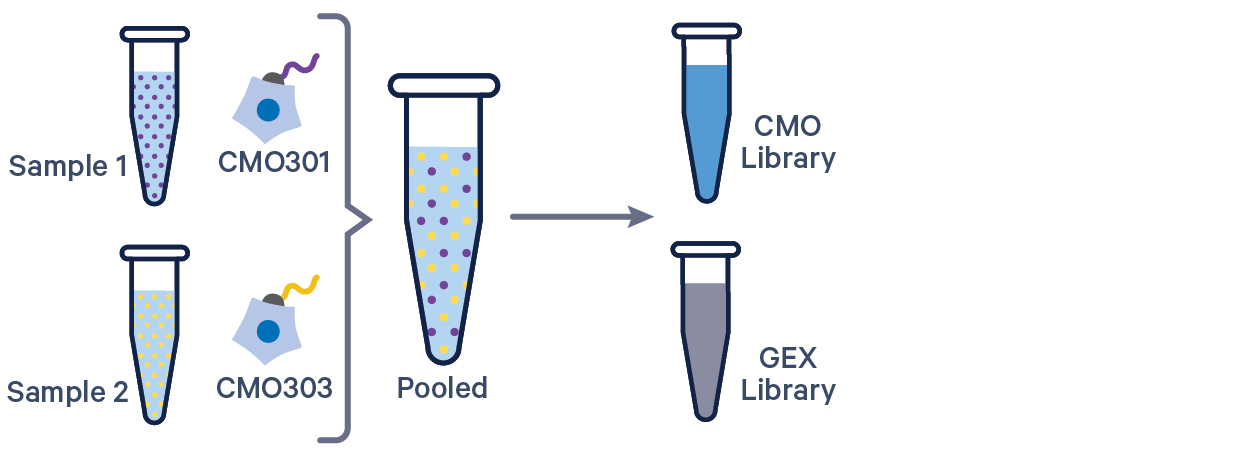 See example dataset |
[gene-expression] reference,/path/to/transcriptome expect-cells, enter expected number of recovered cells include-introns,true |
3' GEX with Cell Multiplexing, multiple CMOs/sample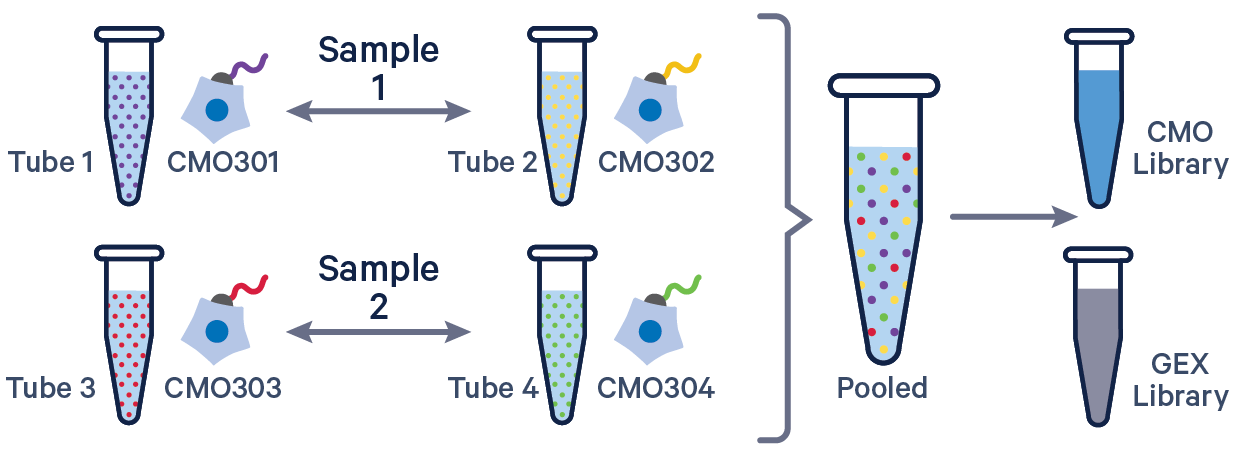 See example dataset. Note usage of the | to separate CMO tags. Learn more about when to use multiple CMOs per sample here. |
[gene-expression] reference,/path/to/transcriptome expect-cells, enter expected number of recovered cells include-introns,true |
3' GEX with Cell Multiplexing and Feature Barcode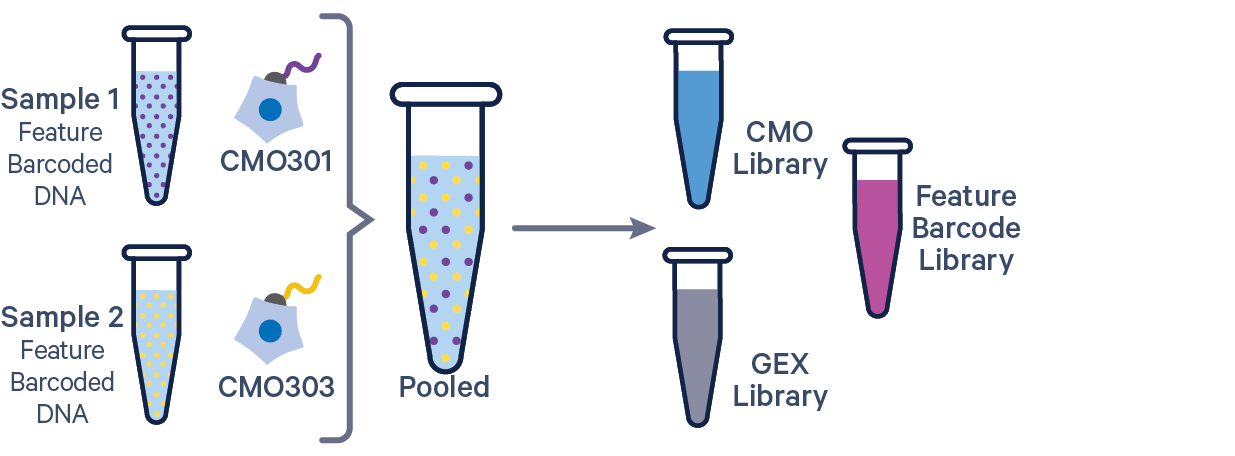 The Feature Barcode in this config CSV example ( [libraries] section) is Antibody Capture. Use CRISPR Guide Capture for CRISPR Feature Barcode experiments. |
[gene-expression] reference,/path/to/transcriptome expect-cells, enter expected number of recovered cells include-introns,true |
The cmo-set option in the [gene-expression] table of the multi config CSV allows you to provide a reference for custom Cell Multiplexing oligos (e.g., antibody TotalSeqA/B/C tags). The design of this reference is nearly identical to the Feature Barcode Reference used to describe Feature Barcodes, with one difference: the feature_type is required to be Multiplexing Capture instead of those feature types supported in the Feature Barcode reference. The id column may contain alphanumeric, underscore, and hyphen characters; special characters like a pipe (|) should not be used in this file (only for separating multiple CMO IDs from the same sample in config CSV).
id,name,read,pattern,sequence,feature_type CMO301,CMO301,R2,5P(BC),ATGAGGAATTCCTGC,Multiplexing Capture CMO302,CMO302,R2,5P(BC),CATGCCAATAGAGCG,Multiplexing Capture CMO303,CMO303,R2,5P(BC),CCGTCGTCCAAGCAT,Multiplexing Capture CMO304,CMO304,R2,5P(BC),AACGTTAATCACTCA,Multiplexing Capture CMO305,CMO305,R2,5P(BC),CGCGATATGGTCGGA,Multiplexing Capture CMO306,CMO306,R2,5P(BC),AAGATGAGGTCTGTG,Multiplexing Capture CMO307,CMO307,R2,5P(BC),AAGCTCGTTGGAAGA,Multiplexing Capture CMO308,CMO308,R2,5P(BC),CGGATTCCACATCAT,Multiplexing Capture CMO309,CMO309,R2,5P(BC),GTTGATCTATAACAG,Multiplexing Capture CMO310,CMO310,R2,5P(BC),GCAGGAGGTATCAAT,Multiplexing Capture CMO311,CMO311,R2,5P(BC),GAATCGTGATTCTTC,Multiplexing Capture CMO312,CMO312,R2,5P(BC),ACATGGTCAACGCTG,Multiplexing Capture
The default CMO reference above is available as a downloadable CSV here.