Cell Ranger7.0, printed on 04/11/2025
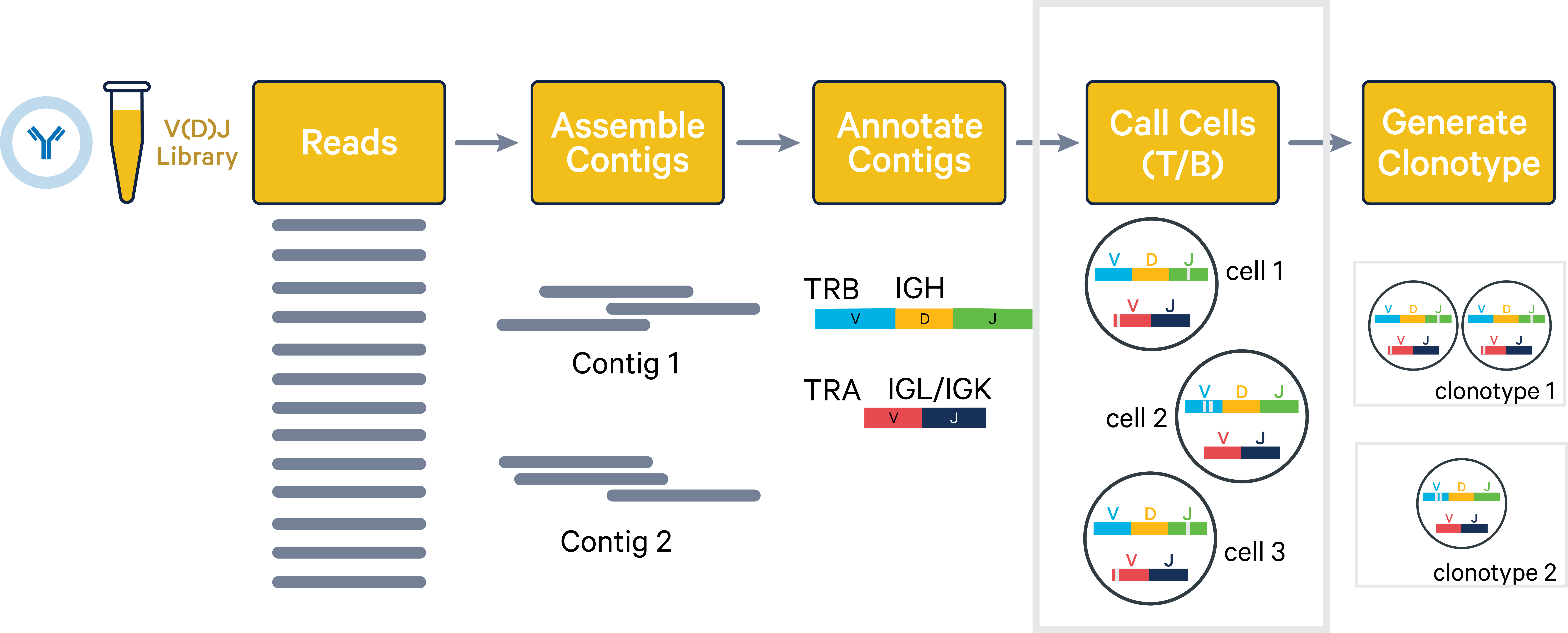
In the 10x Genomics system, a large number of GEMs (Gel Beads-in-emulsion) are generated. A fraction of those GEMs contain a cell, some of which are T or B cell.
T or B cell detection occurs by the identification and counting of V(D)J transcripts from those cells. Some T and B cells have very low expression levels for these transcripts, and thus these may not be detected. Conversely, sufficiently high levels of extracellular mRNA may result in some barcodes being misidentified as T or B cells. Thus the goal of the V(D)J cell calling algorithm is to approximate the set of barcodes that contain a T or B cell. The cell calling algorithm is executed as part of the assembly algorithm.
To be identified as a T or B cell, a barcode must satisfy the following three requirements:
There must be a productive, confident contig. If there is only one such contig, there must be more than one UMI supporting its junction region. In de novo mode, the presence of a contig is the only requirement. Although other cell types can exhibit transcription within the TCR and BCR loci, only T and B cells produce fully rearranged transcripts that contain both a V and a C segment. Therefore, having a productive contig serves as evidence that a transcript from a T or B cell was present in the GEM. However, the transcript may not have arisen from an intact cell (background noise), i.e., captured from fluid between cells. To reduce the likelihood of calling such background transcripts as cells, the algorithm requires each barcode to be supported by more than one UMI.
There must be at least three filtered UMIs having at least two read pairs each (see Assembly Algorithm). This reduces the likelihood of misidentifying a cell as a T or B cell based solely on background transcripts.
Compute the N50 value of the number of read pairs per UMI, across all barcodes. For a given barcode, if the maximum read pair count across filtered UMIs is less than 3% of this N50, do not call the barcode a cell. This provides some protection against transcripts arising from index hopping on an Illumina flowcell, and from other forms of cross-library contamination.
In addition to the three requirements listed above, Cell Ranger v3.1 (and later) has a filter to account for noise introduced by plasma cells and B cells containing large amounts of RNA (as documented in the Cell Ranger 3.1 release notes). This 1) tightens the is_cell filter for low-frequency clones that share a chain with a higher-frequency or large clone, and 2) shrinks high-frequency clones to remove noise from mRNA leakage caused by sample processing (e.g. not due to biological clonal expansion).
Additional cell filters are imposed during clonotype grouping.