Cell Ranger7.0, printed on 04/11/2025
The Chromium Single Cell Immune Profiling Solution with Feature Barcode technology enables analysis of V(D)J clonotypes, gene expression, antibody capture, CRISPR guide capture, and antigen associated capture (Feature Barcode) simultaneously, in the same single cell.
Analysis of each of the sequenced libraries, alone or in combination with other library types, can be performed using the latest versions of Cell Ranger, Loupe Browser, and Loupe V(D)J Browser.
The following illustration depicts how the cellranger multi pipeline can be used to analyze FASTQ data derived from the three Immune Profiling library types from the same GEM well:
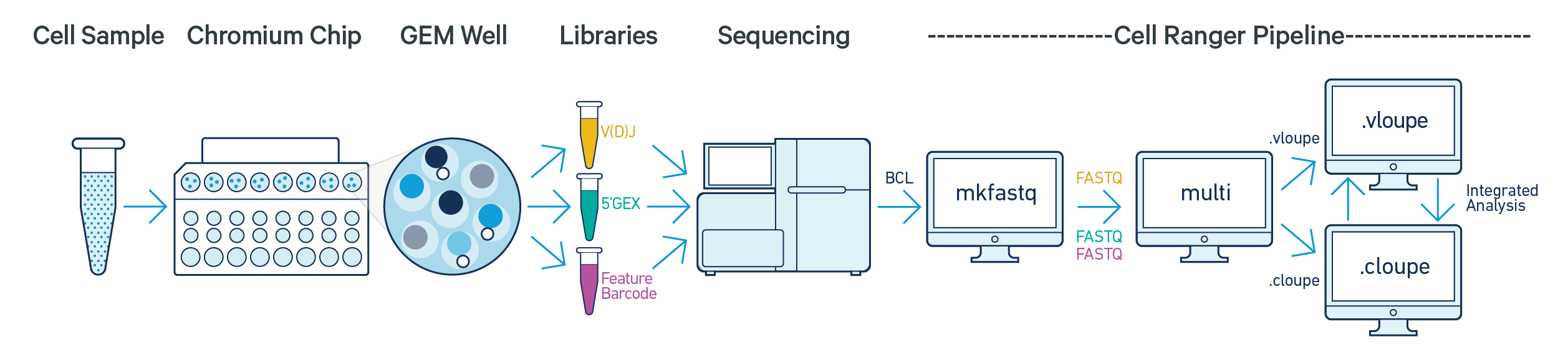
The pipeline will generate a Loupe V(D)J Browser .vloupe file for each of the V(D)J T cell and B cell enriched libraries, in addition to a variety of files to describe V(D)J clonotypes, consensus sequences, and contigs. The pipeline will also generate a Loupe Browser .cloupe file, a feature-barcode matrix, and other information from the Gene Expression and Feature Barcode library. For more details on running the V(D)J analysis pipeline, consult the Cell Ranger multi documentation.
The Loupe Browser and Loupe V(D)J Browser allow you integrate information from the gene expression, cell surface protein (antibody capture), and V(D)J pipeline runs.
To learn more about how to conduct multimodal analysis of V(D)J and Gene Expression data from the same sample, start the Loupe V(D)J + Gene Expression Tutorial. In the tutorial, you will be able to download example 5′ Gene Expression profiles and T cell clonotypes from a non-small cell lung carcinoma, and use the Loupe browsers to analyze the integrated data.
To learn more about analyzing Feature Barcode data, start the Immune Profile Analysis with Feature Barcode and Surface Marker Expression tutorial. In this tutorial, you will download and explore 5′ Gene Expression and Antibody Capture data from a PBMC sample with Loupe Browser.