Cell Ranger7.1, printed on 04/08/2025
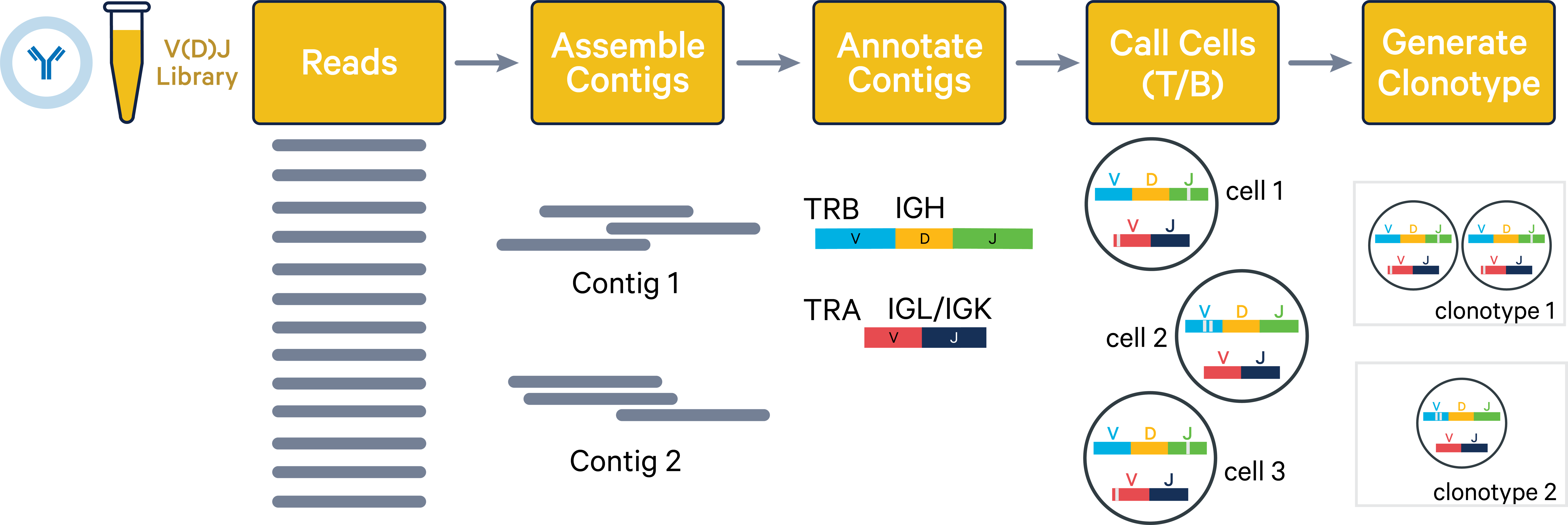
The cellranger vdj pipeline can be used to analyze sequencing data produced from Chromium Next GEM Single Cell 5′ V(D)J libraries. It takes FASTQ files from cellranger mkfastq or bcl2fastq for V(D)J libraries and performs sequence assembly and paired clonotype calling. The pipeline uses the Chromium Cell Barcodes (also called 10x Barcodes) and UMIs to assemble V(D)J transcripts per cell. Clonotypes and CDR3 sequences are output as a .vloupe file which can be loaded into Loupe V(D)J Browser. Visit the What is Cell Ranger page to learn more about Cell Ranger for Immune Profiling.
This page provides an overview of the cellranger vdj algorithm.